Genome‐wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings - Shu - 2019 - Plant Direct - Wiley Online Library

Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho | PNAS
Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains | PLOS Genetics

Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho | PNAS
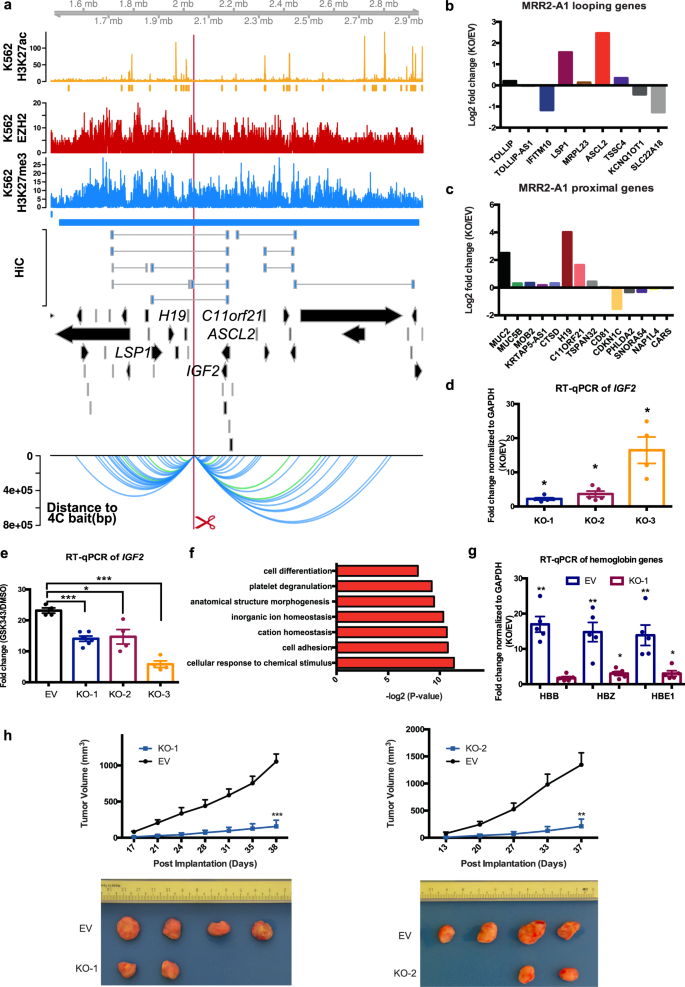
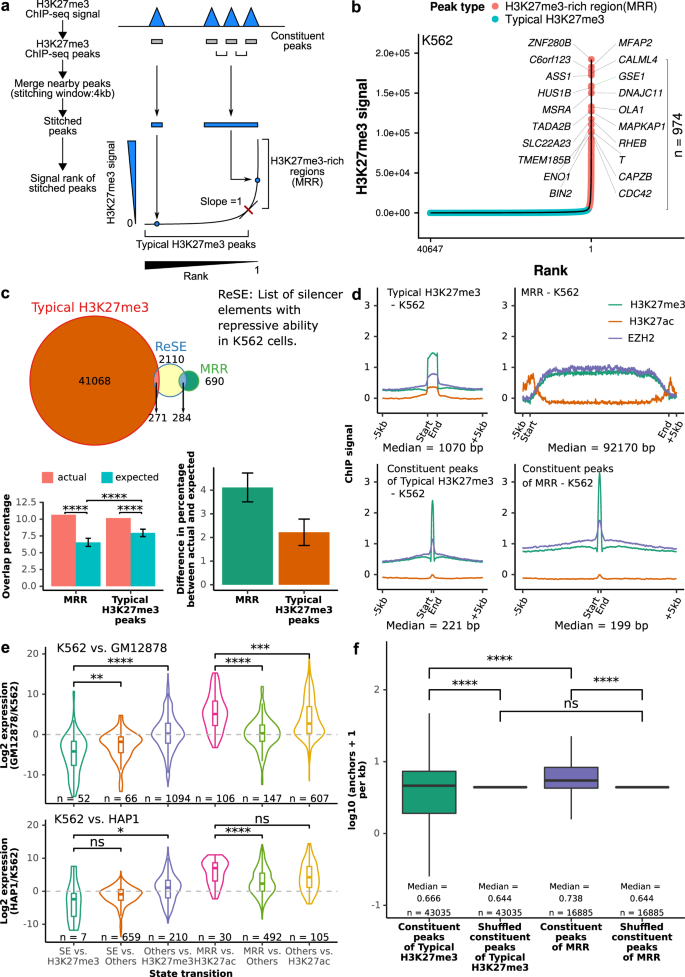
H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions | Nature Communications
Nucleosome Density ChIP-Seq Identifies Distinct Chromatin Modification Signatures Associated with MNase Accessibility

H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions | bioRxiv

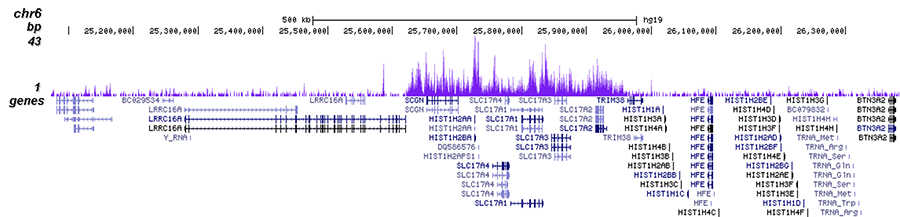
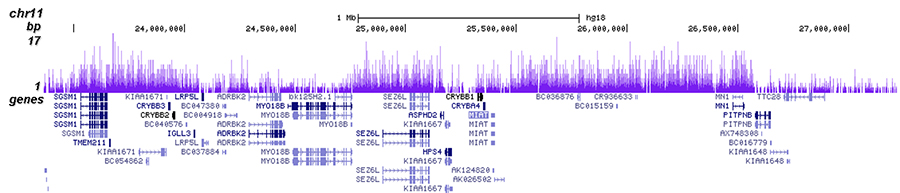
Validation of small-scale ChIP-seq histone mark maps. (A) Two H3K27me3... | Download Scientific Diagram

Figure 2 from ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity | Semantic Scholar

Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho | PNAS
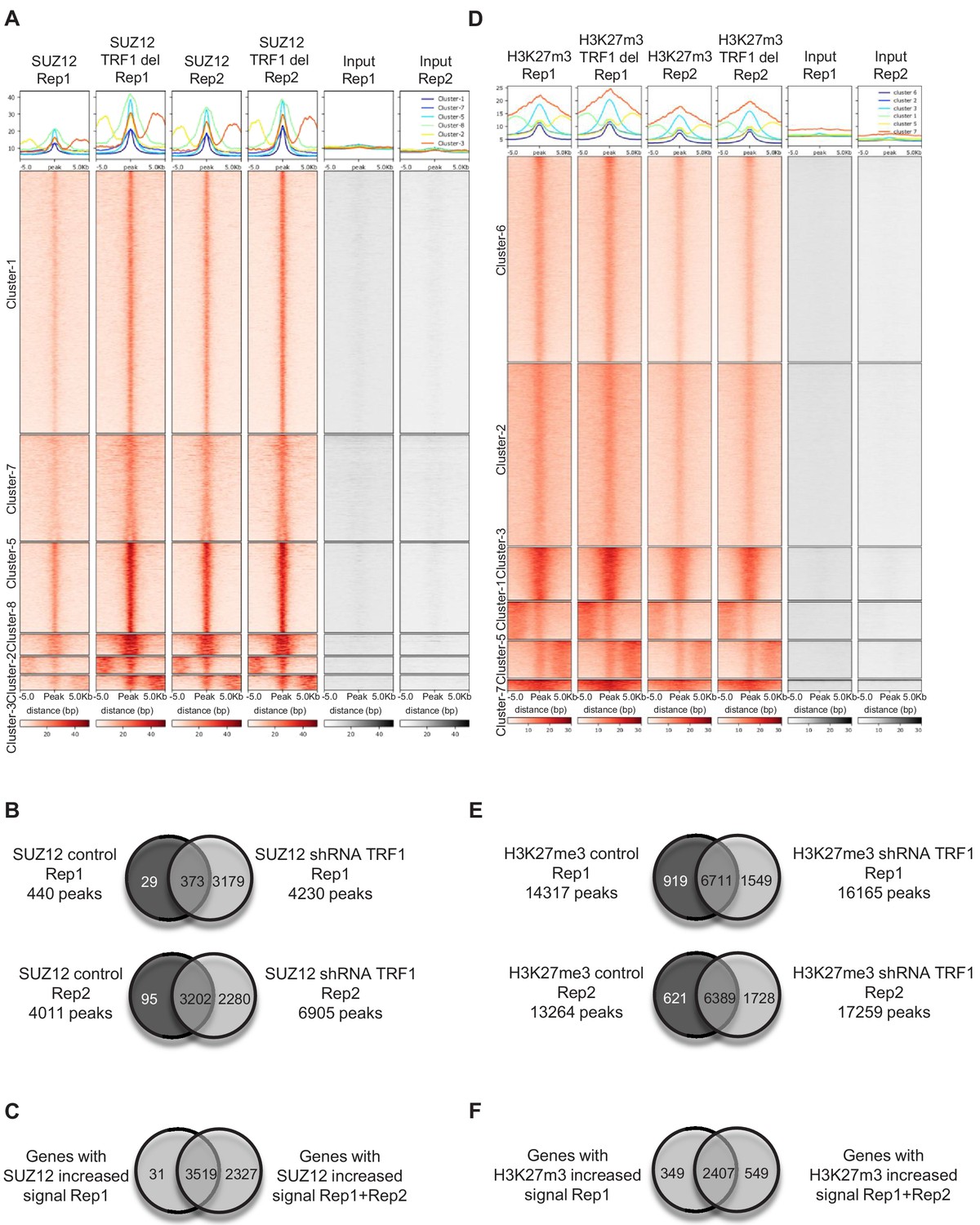
TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2 | eLife

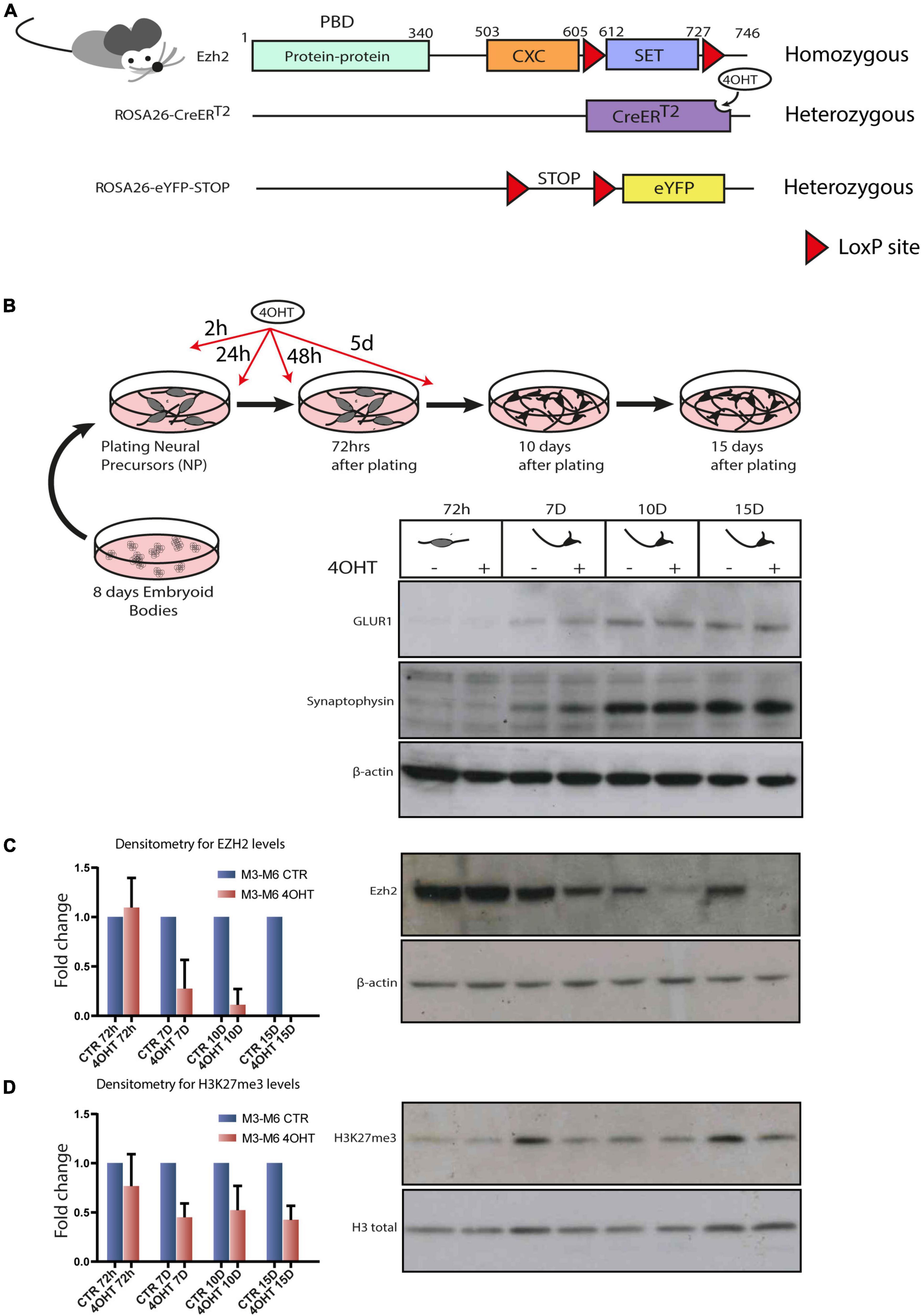
H4K20me1 and H3K27me3 are concurrently loaded onto the inactive X chromosome but dispensable for inducing gene silencing | EMBO reports

ChIP-seq of EZH2 and H3K27me3 in WT iPSCs a, Comparison of the EZH2... | Download Scientific Diagram

H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions | bioRxiv
Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains | PLOS Genetics